JMLS 2022 June;7(1):0-0. 10.23005/ksmls.2022.7.1.52 Epub 2022 June 19
Copyright © 2022 by The Korean Society of Marine Life Science
Analysis of Complete Mitochondrial Genomes of Three Gadus chalcogrammus Specimens (Teleostei; Gadiformes; Gadidae) from Korea and Japan
Chung Il Lee; Department of Marine Bioscience, Gangneung-Wonju National University, Gangneung 25457, Korea
Hae Kun Jung; East Sea Fisheries Research Institute, National Institute of Fisheries Science, Gangneung 25407, Korea
Hae-Kyun Yoo; East Sea Fisheries Research Institute, National Institute of Fisheries Science, Gangneung 25407, Korea
Hyun-Woo Kim; Department of Marine Biology, Pukyong National University, Busan 48513, Korea
Hyun Je Park; Department of Marine Bioscience, Gangneung-Wonju National University, Gangneung 25457, Korea
Chang-Keun Kang; School of Earth Sciences & Environmental Engineering, Gwangju Institute of Science and Technology, Gwangju 61005, Korea
Jeong Hee Shim; East Sea Fisheries Research Institute, National Institute of Fisheries Science, Gangneung 25407, Korea
Keun-Yong Kim; AquaGenTech Co., Ltd., Busan 48300, Korea
Joo Myun Park; Dokdo Research Center, East Sea Research Institute, Korea Institute of Ocean Science & Technology, Uljin 36315, Korea
Moongeun Yoon; National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea
- Abstract
Mitochondrial genomes of three specimens of Gadus chalcogrammus Pallas 1,814 from Korea and Japan were completely analyzed by the primer walking method. They were 16,570~16,571 bp in length, each comprising 13 protein-coding genes, two ribosomal RNA genes and 22 transfer RNA genes. Their gene orders were identical to those of conspecific specimens, but exhibited unique haplotypes. In the phylogenetic tree, the juvenile Korean and adult Japanese specimens were separated from the dominant clade composed of specimens from Japan, Korea, the Bering Sea, and the Arctic, including the adult Korean specimen.
Keywords: Gadus chalcogrammus Japan Korea Mitochondrial genome Phylogeny
Correspondence to: Moongeun Yoon; National Marine Biodiversity Institute of Korea, Seocheon 33662, Korea; E-mail: mgyoon@mabik.re.kr
- Received
- 23 March 2022;
- Revised
- 29 March 2022;
- Accepted
- 12 April 2022.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
The walleye pollock, Gadus chalcogrammus Pallas 1,814 (Teleostei; Gadiformes; Gadidae) is a demersal fish widely distributed in the North Pacific Ocean (Froese and Pauly, 2019). The species lives between depths of 50~500 m and forms an intermediate key component of the marine food web as both a predator and prey items (Funamoto, 2018). Although its population has dramatically declined in the last two decades, uncertainty in its genetic in- formation limits our understanding of its population structure.
Three specimens of G. chalcogrammus from Korea, of different ages (juvenile and adult), and Japan (adult) were collected at Goesong in Korea (from 38°18'00"N, 128°35'00"E) and Otobe in Japan (41°57'44"N, 139°50'47"E). The voucher specimens were deposited at the aquatic animal collection of the Department of Marine Bioscience, Gangneung-Wonju National University (Gangneung, South Korea) under voucher numbers, KGB01, KGC01, and JO01. Genomic DNA was extracted from fin tissues according to Asahida et al. (1996) and deposited at the aquatic animal collection of the Department of Marine Bioscience. Their meta- genomes were amplified through two independent and over- lapping PCR runs, and the PCR products were directly sequenced using a set of 39~41 primers. The sequence data that support the findings of this study are openly available in GenBank (https:// www.ncbi.nlm.nih.gov/) under the accession numbers MW288641- MW288643.
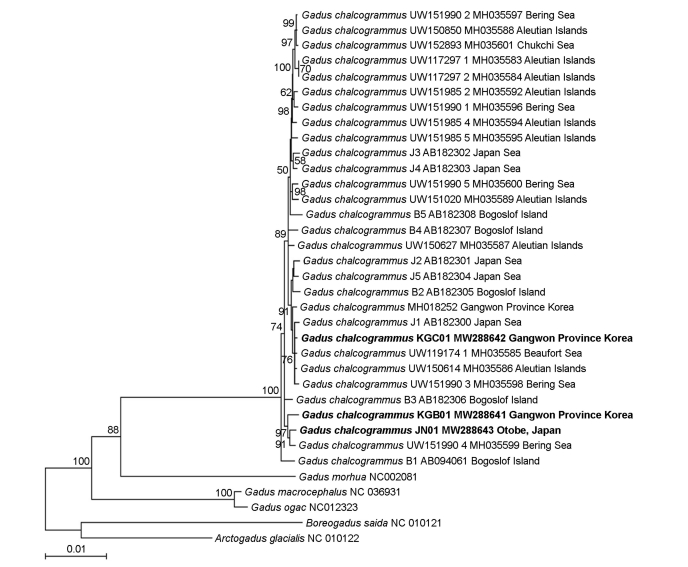
All complete mitogenome sequences of 27 specimens of G. chalcogrammus were retrieved from GenBank. They were aligned and refined manually to correct obvious misalignments. The nucleotide matrix of 13 protein-coding genes (PCGs) (11,410 bp), two ribosomal RNA (rRNA) genes (2,623 bp), and 22 transfer RNA (tRNA) genes (1,536 bp) excluding non-coding regions were used to reconstruct a phylogenetic tree with MEGA7 (Kumar et al., 2016) for maximum likelihood (ML) analysis. Tree support was evaluated by bootstrapping (1,000 replications).
The three complete mitogenomes of G. chalcogrammus in this study were circular molecules of 16,570~16,571 bp in length, consisting of 13 PCGs, two rRNA genes, and 22 tRNA genes. Their gene orders were identical not only to conspecific specimens (Yanagimoto et al., 2004; Sim et al., 2018) but also to those of typical vertebrates. They exhibited unique haplotypes by showing genetic distance of 0.0008-0.0048 among all G. chalcogrammus available in GenBank across all mitogenome sequences.
In the ML tree, Gadus species formed a strongly supported monophyletic group, and the three specimens from Korea and Japan formed a monophyletic group with conspecific specimens in GenBank (Fig. 1). Within the G. chalcogrammus lineage, the juvenile Korean (KGB01) and adult Japanese (JN01) specimens formed a distinct clade with one of conspecific specimens in GenBank and separated from the dominant clade composed of specimens from Japan, Korea, and the Bering Sea, and the Arctic, including the adult Korean specimen (KGC01). The mitogenome information in this study will provide baseline data for population structure, effective management, and recovery plan of a valuable fisheries resource, G. chalcogrammus.
- References
-
3. Froese R, Pauly, D. 2019. FishBase. World Wide Web electronic pub- lication. [accessed 2020 October 8]. http://www.fishbase.org/
-
6. Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolu- tionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874. https://doi.org/10.1093/molbev/ msw054